Quantification of protein interactions and solution transport using high-density GMR sensor arrays
- Source: [ @gaster2011 Quantification of protein interactions and solution transport using high-density GMR sensor arrays ]
- Tags: #giant-magnetic-resistance #biosensor #protein-dynamics
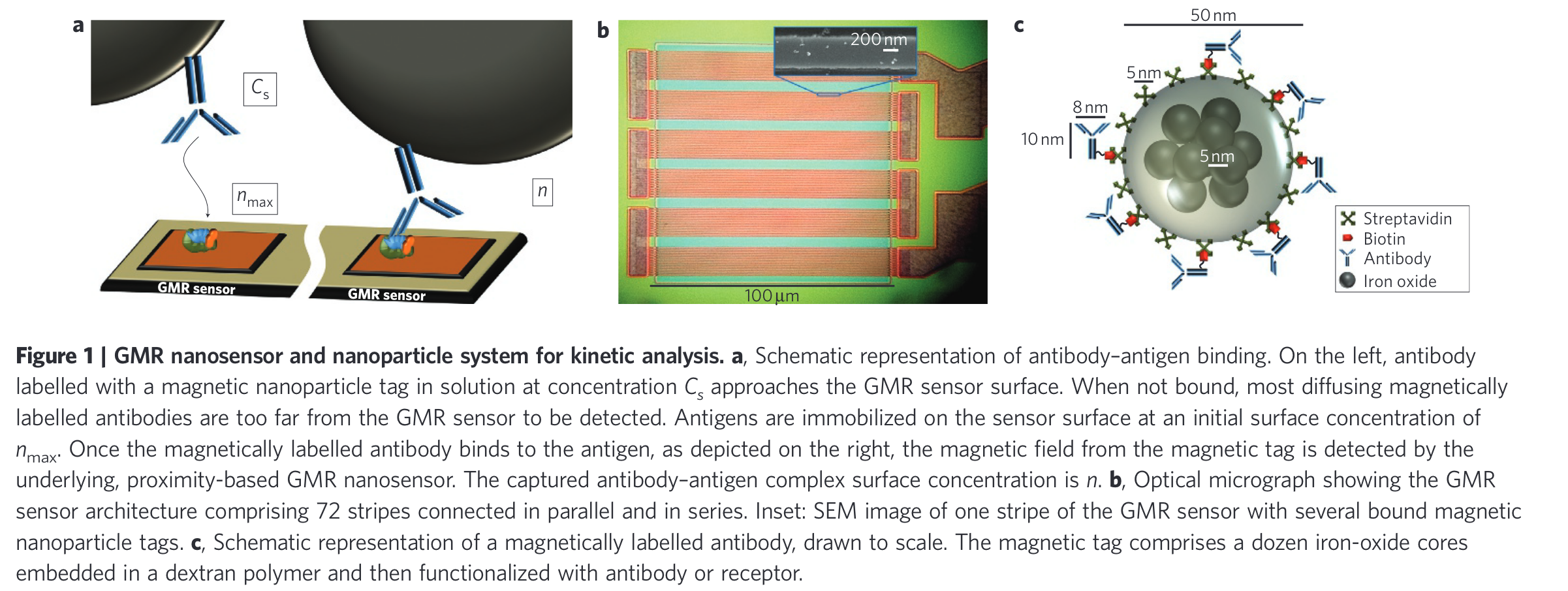
Studying protein dynamics is crucial for many aspects of biology, from DNA hybridization, antigen-antibody binding, and DNA-protein interaction. However, one of the challenges is that the introduction of labels alter the behavior of the studied object (for instance, the diffusion coefficient changes, etc.)
The paper by [ @gaster2011 Quantification of protein interactions and solution transport using high-density GMR sensor arrays ] focuses on using magnetic nanoparticles in combination with giant magnetic resistance sensors. A functionalized surface can trap the magnetic beads and their presence can be detected in parallel, in real-time .
The core message of the work is the development of a model to characterize the protein dynamics between labeled objects and a surface. The model is semi-analytical and seems to describe properly the detected signals.
The biggest advantage over surface plasmon resonance sensors is the large dynamic range (6 log or more compared to 2 log for SPR), and the concentration:
This work opens the door to profiling the affinity of specific compounds against an entire proteome.
There is a spinout associated with the project called Flux Biosciences
Backlinks
These are the other notes that link to this one.
