Literature/202306221512 sers to identify cancer evs
It is an interesting paper, in which they show that surface enhanced raman spectroscopy can be used to classify samples of extracellular vesicles belonging to cancer or healthy patients.
They prepare some slides by depositing
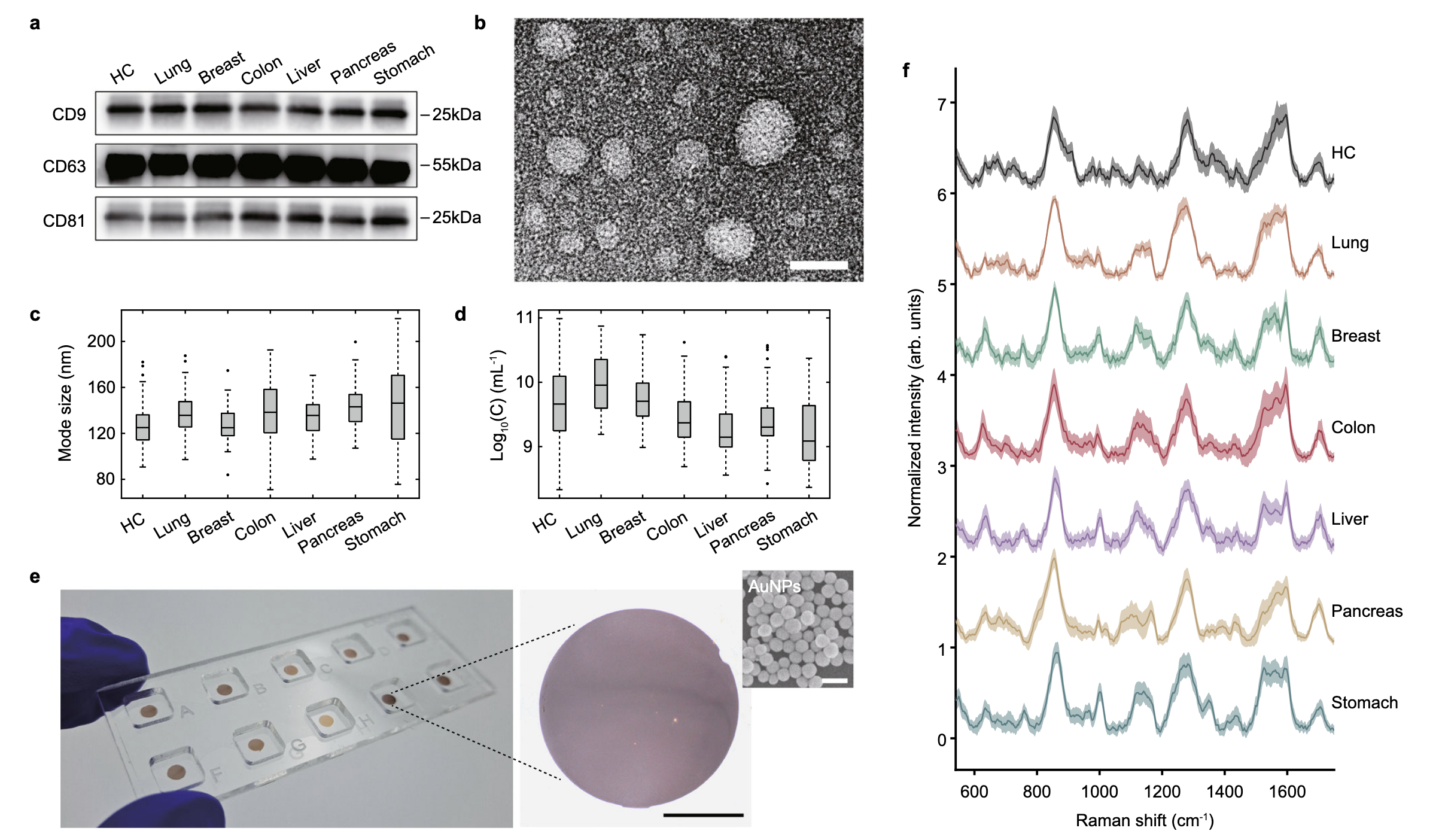
The image above shows an example of the slides they prepare and the spectra they acquired. They feed the spectra to a machine-learning classifier and obtain around a 96% accuracy. They can also push forward and use the same approach to identify the tissue of origin (TOO).
However, I am very skeptical about the approach:
- The particles are very big compared to the exosomes, but that may still be OK.
- The gap in which the SERS happens is too small to fit an entire vesicle, therefore:
- What are the odds that the specific differentiating protein is sitting directly in the hotspot?
- The authors have analyzed "100 signals" per sample. I guess that means they recorded spectra in 100 different locations. How many EV's can there by in each one of those spots? With a focal spot of around 500nm, that gives a very small number of AuNP, so assuming around 10 EV's per spot, they authors are analyzing only 1000 EV's per sample.
I don't think there is any way in which such a small number of EV's can be enough to get the few ones that may be coming from cancerous tissue, unless the sample was enriched to levels never heard of.
The fact that they can train a machine learning algorithm under those conditions, raises many questions. The data generated should not allow to train any algorithm to a statistical significance, but the authors managed to get 96% accuracy.
It is still too small of population screening purposes, but a surprising number all things considered.
Backlinks
These are the other notes that link to this one.
