Proteomeic profile of evs
Since it is challenging to distinguish HIV viruses from EVs simply by using biomarkers or physical properties (see: HIV-1 contains CD9 and CD63), an alternative approach is to study the proteomic profile of the particles[@martinjaular2021Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection] that can be harvested from cell cultured medium.
It is important to note several things, first that Tetraspanins are unevenly distributed across extracellular vesicles, therefore they can't be used as a single indicator (regardless of the method). Moreover, the abundance of tetraspanins in EVs can be linked to the sample preparation protocol, but this is not discussed in depth.
Therefore, a better approach is to study cross-correlations between the presence of proteins, which would indicate some affinity between them.
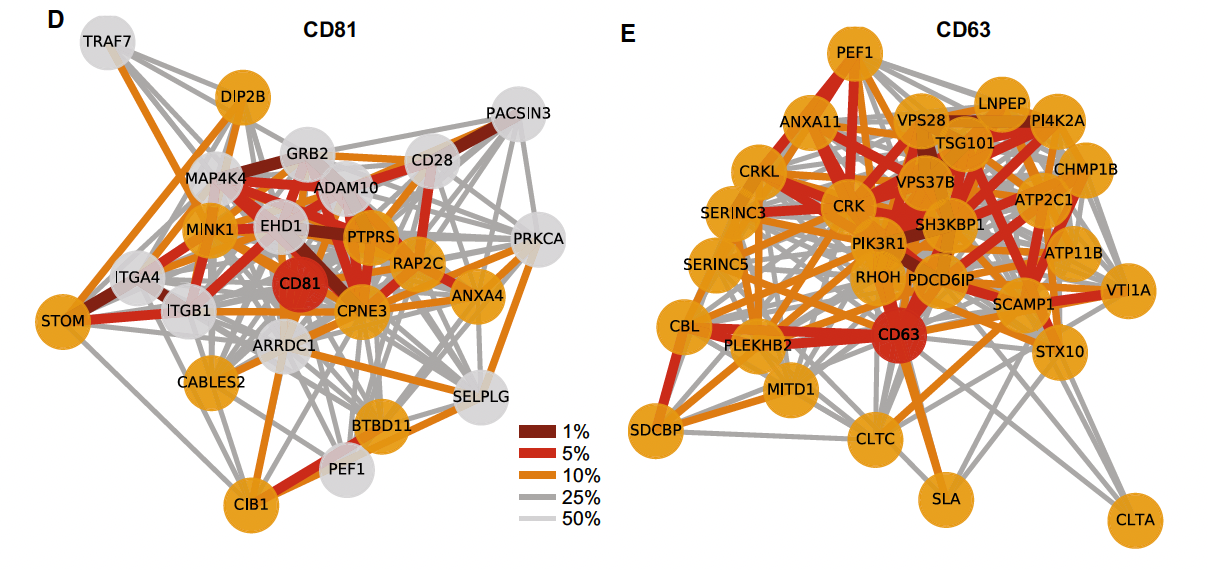
The figure above shows a network plot of the likelihood of finding other proteins given a protein of interest, in this particular case, CD81 and CD63. This is particularly interesting since it can be used to determine the probability of finding two proteins given some initial conditions.
Besides the application to study HIV shown in [@martinjaular2021Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection], this kind of databases can be incredibly useful, and it is available online.
Backlinks
These are the other notes that link to this one.
