Literature/202301141333 comparing simoa and sp-iris
- Source: [@frigerio2022Comparing digital detection platforms in high sensitivity immune-phenotyping of extracellular vesicles]
- Tags: #SP-IRIS #SIMOA #digital-detection #Extracellular-Vesicle
Simoa and SP-IRIS are used and compared in their abilities to detect extracellular vesicles. Both are digital methods (see: Analog versus digital detection of molecules). And they can be a crucial tool to detect and quantify Low abundance EVs in bio-fluid derived samples.
Using the SiMoA platform
The SiMoA platform was from Quanterix, and they used the Homebrew Kit (I assume a kit that allows custom bead functionalization). The concentrations of beads used was
Using the SP-IRIS platform
The NanoView Biosciences instrument was used, with custom silicon chips. Chips were incubated for 2 hours, and then placed on a shaker for 3min. Fluorescent antibodies were added and the chips were incubated for another hour.
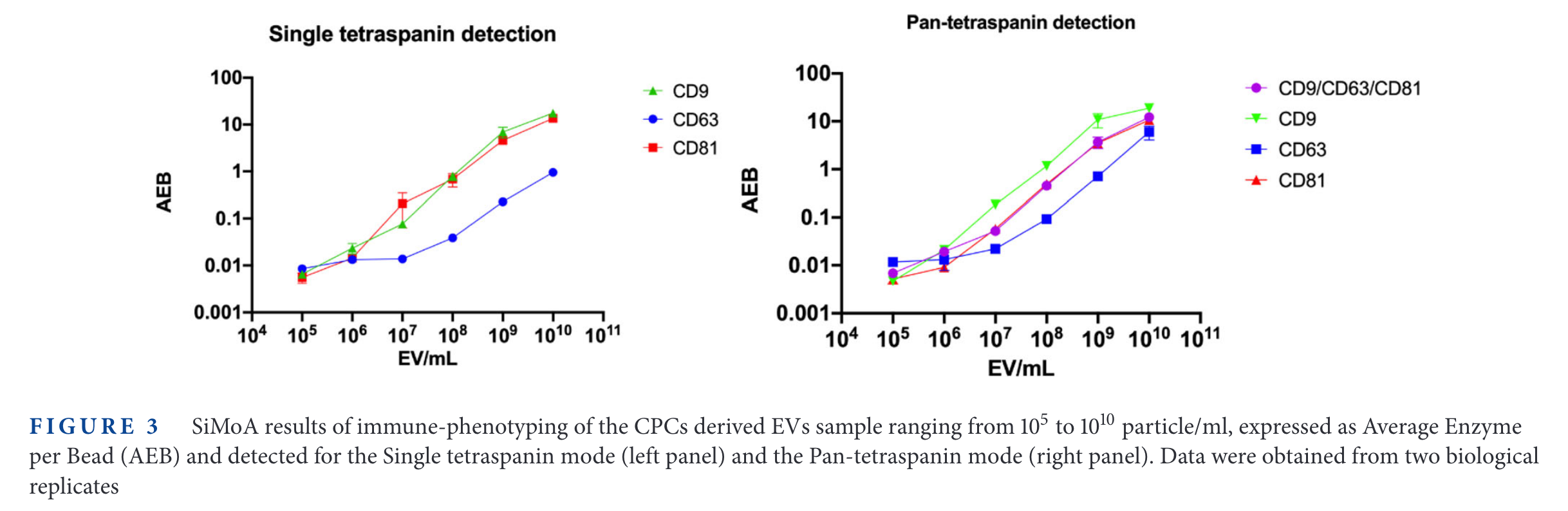
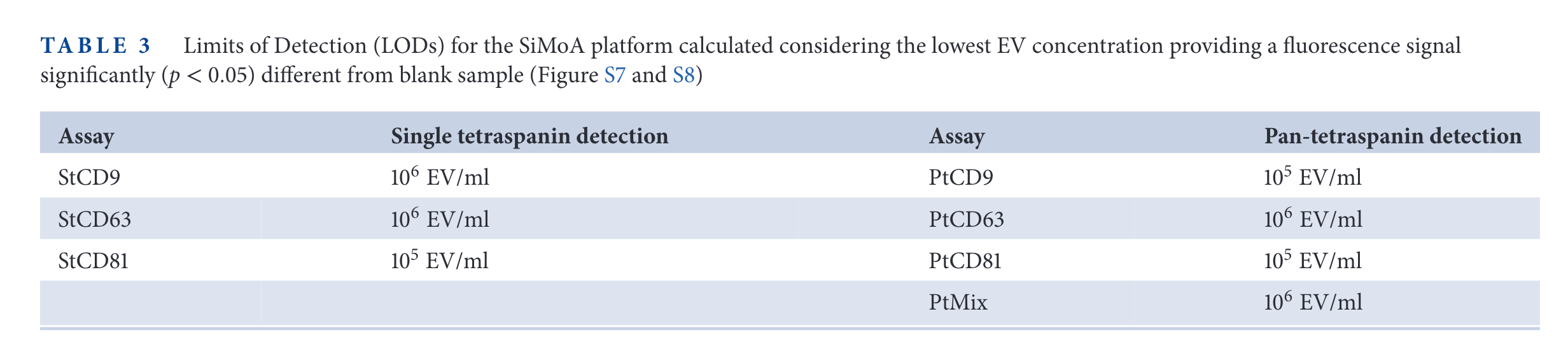
SiMoA results
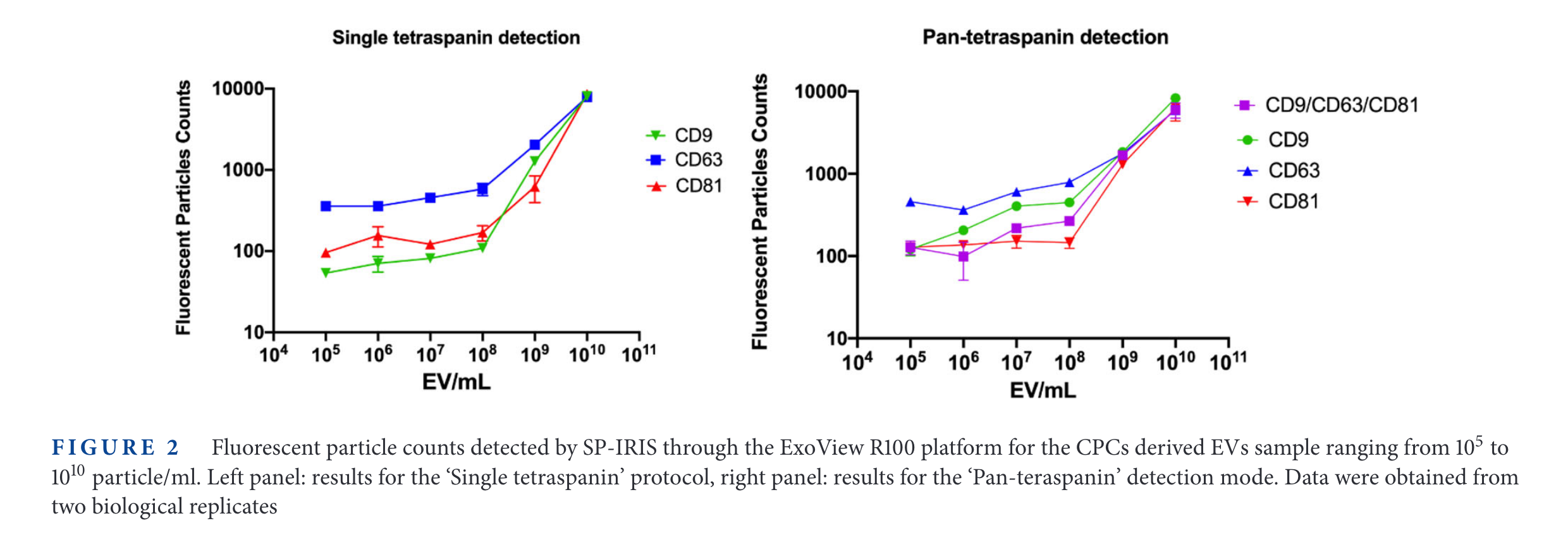
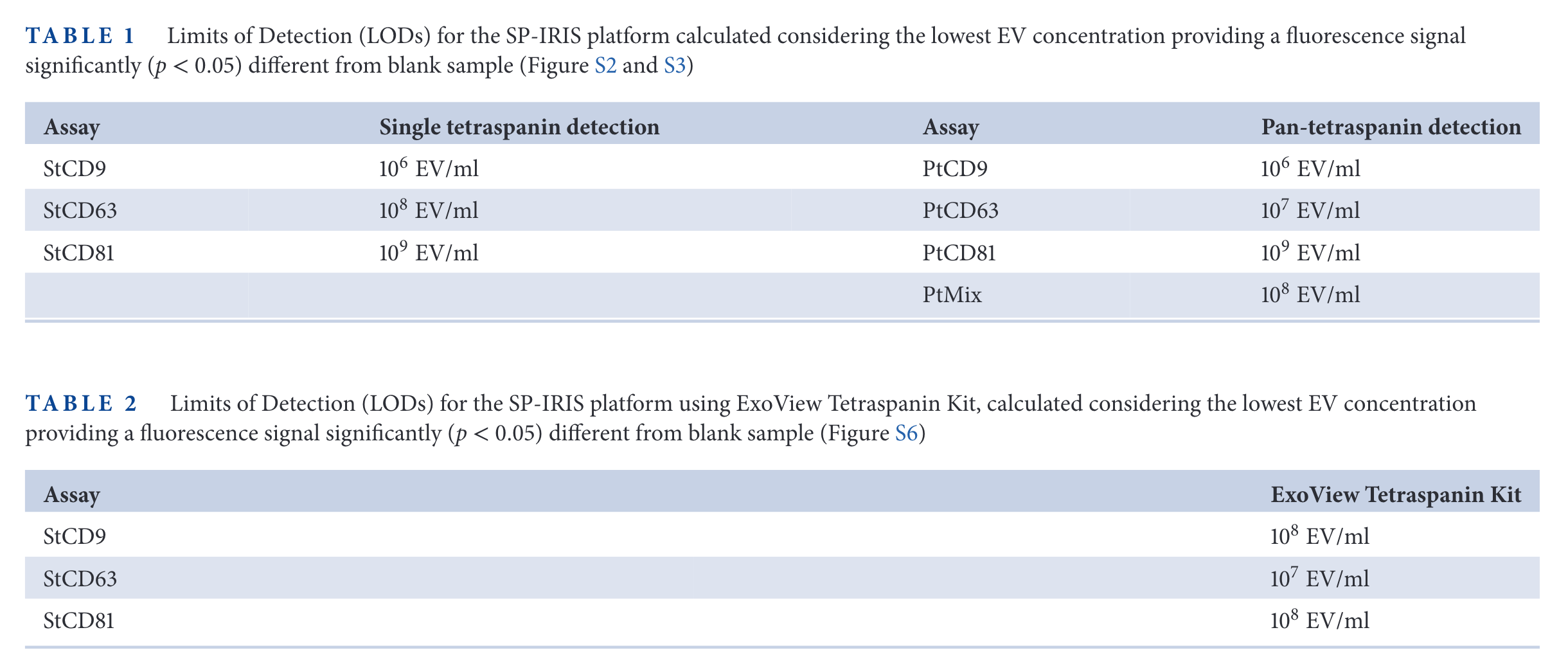
SP-IRIS results
Comparison on same cell-samples
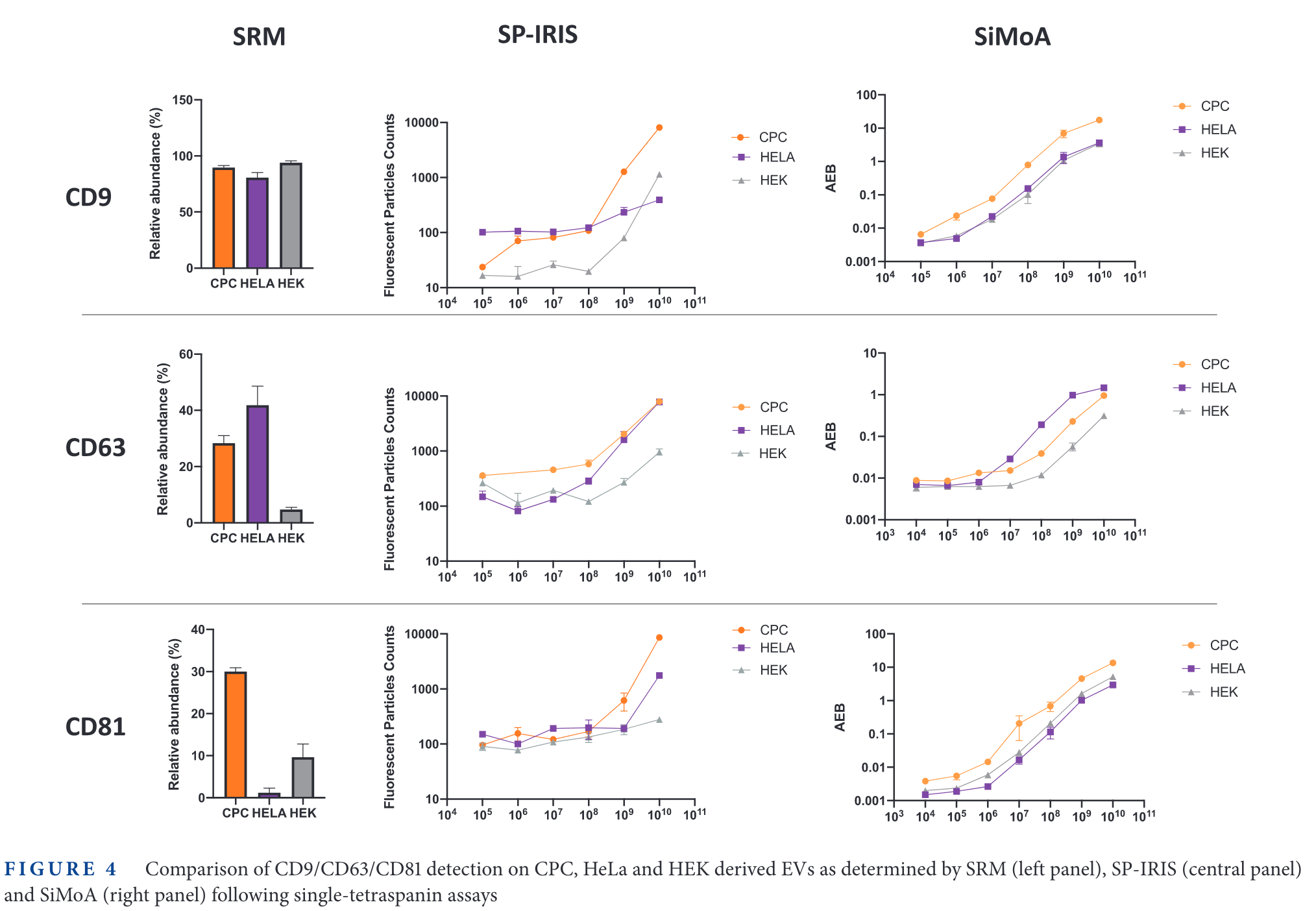
In the figure above, the relative expression of some tetraspanins for different cell lines is measured. Then, SP-IRIS and Simoa are used to measure those same proteins. The limit of detection of SP-IRIS is worse than that of SiMoA for low abundance specimens (CD63 in HEK cells, CD81 in HeLa cells, for examples).
It is interesting that the 'background' signal changes depending with the tetraspanin targetted. For example, in CD9, the 'Fluorescence Particle Counts' of SP-IRIS goes down to ~10, while for CD81 is always above 100. On the other hand, the SiMoA instrument goes from 0.001 in CD9 and CD81 up to 0.01 in the case of CD63.
Backlinks
These are the other notes that link to this one.
