Simoa
Simoa stands for single-molecule arrays[@rissin2010Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations] and essentially consists of an essay to detect very small concentrations of antigens, following protocols very similar to those of the Elisa test.
The general principle is like in the image below:
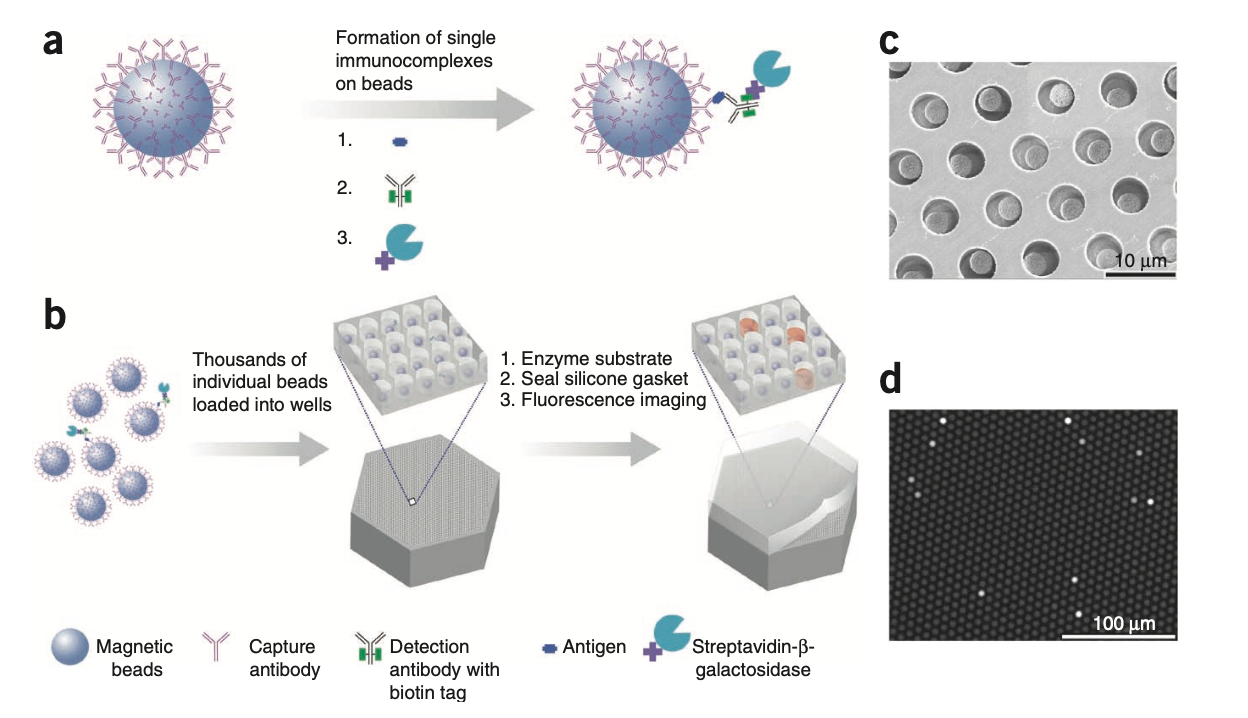
Paramagnetic beads, coated with a known antibody can diffuse in the medium, capturing antigens. The process is the same as in the Sandwich ELISA (see: Elisa test). They are later precipitated onto specific arrays in which only one bead fits.
By looking at each well with and without the beads and with or without fluorescence, it is possible to conduct a digital elisa test.
The method can detect as little as $10^{-19}M$ under special conditions, or $10^{-15}M = 10fM$ in clinical conditions. See: why pushing sensitivity of assays.
One of the core aspects of the Simoa devices is how to know if you have double binding events. In [@rissin2010Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations], the authors present a statistical argument in which double binding events is almost negligible if binding (overall) is below 50%, as it follows a poissonian distribution.
See also: Tyranny of the Langmuir binding isotherm
Backlinks
These are the other notes that link to this one.
